Stretching out Support Vector Machine with Calibrated Estimation
Data:
3000 lines sentiment text data with labels 0 or 1
Work flow: (explaination to the methods will be included in each steps)
- Text preprocessing
- Text cleaning
- Tokenizing
- Vectorizing
- Fitting Support vector machine(run cross-validation to select suitable slack variable)
- Calibration, learn a monotonic transformation by 3 methods
- sigmoid
- platt scaling
- isotonic regression
- Visualizing result and Conclusion
- import modules
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from string import digits, punctuation
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score
from sklearn.svm import LinearSVC
from sklearn.model_selection import GridSearchCV
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.isotonic import IsotonicRegression
from sklearn.calibration import calibration_curve
%matplotlib inline
plt.style.use('seaborn-whitegrid')
Step 1: Text preprocessing
We will use the sentiment data which contains 3000 lines, each with one sentence followed by a label (0 or 1).
s = pd.read_csv('full_set.txt', header=None, sep='\t', names=['sentence', 'label'])
- remove the labels from the sentences and store them separately.
label = s.pop('label')
y = label.values
- remove digits and punctuation, and make everything lowercase.
# remove punctuation
s['sentence'] = s['sentence'].apply(lambda x: x.translate(str.maketrans('', '', punctuation)))
# remove digits
s['sentence'] = s['sentence'].apply(lambda x: x.translate(str.maketrans('', '', digits)))
# lowercase strings
s['sentence'] = s['sentence'].apply(lambda x: x.lower())
- remove “stop words”, common words that are useless for classification.
stopwords = ['the','a','an','i','he','she','they','to','of','it','from']
s['sentence'] = s['sentence'].apply(\
lambda x: ' '.join([word for word in x.split() if word not in (stopwords)]))
# check text after preprocessing
s
output:
| sentence | |
|---|---|
| 0 | so there is no way for me plug in here in us u... |
| 1 | good case excellent value |
| 2 | great for jawbone |
| 3 | tied charger for conversations lasting more th... |
| 4 | mic is great |
| ... | ... |
| 2743 | just got bored watching jessice lange take her... |
| 2744 | unfortunately any virtue in this films product... |
| 2745 | in word is embarrassing |
| 2746 | exceptionally bad |
| 2747 | all in all its insult ones intelligence and hu... |
2748 rows × 1 columns
- vectorizing: Bag-of-words
corpus = s['sentence'].values
vectorizer = CountVectorizer(max_features=5000)
X = vectorizer.fit_transform(corpus).toarray()
# add 1 to the end of each row to represente intercepts
b = np.ones((X.shape[0], 1))
X_b = np.hstack((X, b))
# check output
X_b.shape
output:
(2748, 5001)
- spliting data
Divide the 3000 data points into a training set of size 2200, a calibration set of size 400, and a test set of size 400. Keep the classes balanced (each set should be evenly split between positives and negatives).
X_train, X_, y_train, y_ = train_test_split(X_b, y, test_size=4/15, stratify=y, random_state=42)
X_calib, X_test, y_calib, y_test = train_test_split(X_, y_, test_size=0.5, stratify=y_, random_state=42)
print(f"Dimension of X_train: {X_train.shape}")
print(f"Dimension of y_train: {y_train.shape}")
print(f"Dimension of X_calib: {X_calib.shape}")
print(f"Dimension of y_calib: {y_calib.shape}")
print(f"Dimension of X_test: {X_test.shape}")
print(f"Dimension of y_test: {y_test.shape}")
output:
Dimension of X_train: (2015, 5001)
Dimension of y_train: (2015,)
Dimension of X_calib: (366, 5001)
Dimension of y_calib: (366,)
Dimension of X_test: (367, 5001)
Dimension of y_test: (367,)
Step 2: Fitting Support vector machine(cross-validation to select suitable slack variable)
def cross_valid(C):
score = []
for c in C:
svc = LinearSVC(C=c, max_iter=10000)
clf = svc.fit(X_train, y_train)
score.append(1-np.mean(cross_val_score(clf, X_train, y_train, cv=5)))
return score
C = [0.001, 0.01, 0.1, 1, 10, 100]
err = cross_valid(C)
fig = plt.figure(figsize=(12, 6))
plt.plot(C, err, '-ok', color='slateblue')
plt.xscale('log')
plt.title('Test Error with different C', fontsize=16)
plt.xlabel('C values (log)', fontsize=16)
plt.ylabel('Test Error', fontsize=16);
output:
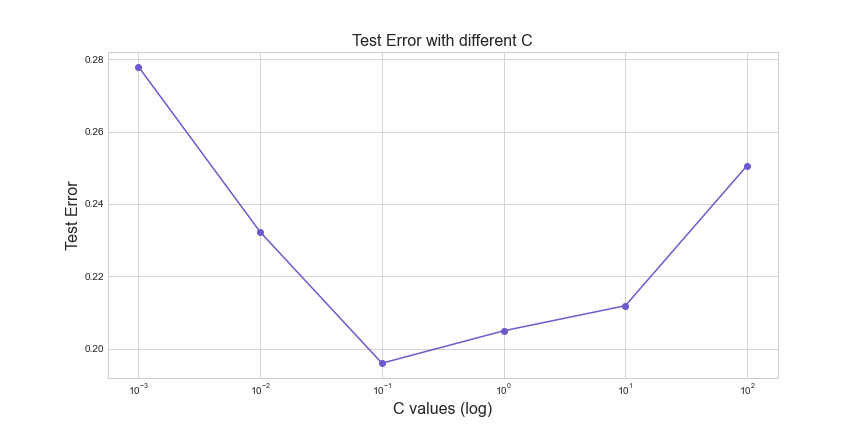
Best C is 0.1 for training set.
svc = LinearSVC(max_iter=10000, C=0.1).fit(X_train, y_train)
print(f"Error rate on test set: {1-svc.score(X_test, y_test)}")
output:
Error rate on test set: 0.16893732970027253
Step 3: Learning a monotonic transformation
We now have a fixed classifier: the label for \(x\) is \(sign(w \dot x + b)\). We will think of the score on point \(x\) as \(z(x) = w \dot x + b\). We want to convert these real-valued scores into probabilities, using a monotonically increasing function \(f\): we will predict \(Pr(y = 1|x) = f(z(x))\). We will investigate three choices for \(f\).
The squashing function
This involves no learning: map score \(z\) to probability \(1/(1+e^{-z})\)
Platt scaling
We will map score \(z\) to probability \(1/(1+e^{-(az+b)})\). Hence \(a,b \in \mathbb{R}\) are parameters to be learned using the calibration set, using maximum-likelihood. That is, we want to find \(a, b\) to minimize
\[\sum_{(x,y)\in C} \ln(1+exp(-y(az(x)+b)))\]where \(C\) is the calibration set, assuming labels are \(±1\). Since \(z(x)\in R\), this is essentially a one-dimensional logistic regression problem, with data \(\{(z(x),y):(x,y) \in C\}\), and can be solved easily, in 1-2 lines, using sklearn.linear_model.LogisticRegression.
Isotonic regression
The idea is to learn a general monotonic function \(f : \mathbb{R} -> [0, 1]\) from scores to probabilities. The data will be \(\{(z(x), y) : (x, y) \in C\}\), assuming the labels are \(y \in \{0, 1\}\) (not \(±1\)). Suppose \(Z\) is a vector of \(z\)-values of the calibration set, and \(Y\) is a vector of corresponding 0-1 labels. (Both are \(400\)-dimensional vectors.) To learn the monotonic transform:
from sklearn.isotonic import IsotonicRegression
isotonic_clf = IsotonicRegression(out_of_bounds=’clip’)
isotonic_clf.fit(Z,Y)
To get probabilities for points in the test set, let \(Z^*\) be the vector of test set scores. Use:
test_probs = isotonic_clf.transform(Z_star)
class CalibratedEstimator():
def __init__(self, clf, option, forcing=False):
self.clf = clf
self.option = option # which option to calibrate
self.forcing = forcing # forcing intercept b = 0 or not
def __z_score(self, X):
mean = np.mean(X)
stdev = np.std(X)
return (X - mean)/stdev
def __z(self, X, w):
return np.dot(X, w.T)
def __sigmoid(self, x):
return 1/(1+np.exp(-x))
def __platt_scaling(self, X, a, b):
return 1/(1+np.exp(-(a*X + b)))
def pre_train(self, X_train, y_train):
"""Calculate coefficients trained by pre-defined classifier
"""
if self.forcing == True:
X_train = X_train[:, :-1] # ensure intercepts are 0
self.clf.fit(X_train, y_train)
self.w = self.clf.coef_
return self
def fit(self, X_calib, y_calib):
"""Fit monotonic transformation with options
"""
if self.forcing == True:
X_calib = X_calib[:, :-1]
z_calib = self.__z(X_calib, self.w)
if self.option == 'platt_scaling':
lr = LogisticRegression().fit(z_calib, y_calib)
self.a, self.b = lr.coef_, lr.intercept_
elif self.option == 'isotonic':
Z_c = self.__z_score(z_calib) # calculate Z-score (calib set)
Z_c = Z_c.reshape((Z_c.shape[0],))
self.iso = IsotonicRegression(out_of_bounds='clip').fit(Z_c, y_calib)
return self
def predict_proba(self, X_test):
"""Predict confidence score based on calibrated coefficients
"""
if self.forcing == True:
X_test = X_test[:, :-1]
z_test = self.__z(X_test, self.w)
if self.option == 'sigmoid':
prob_pred = self.__sigmoid(z_test)
elif self.option == 'platt_scaling':
prob_pred = self.__platt_scaling(z_test, self.a, self.b)
elif self.option == 'isotonic':
Z_t = self.__z_score(z_test) # calculate Z-score (test set)
Z_t = Z_t.reshape((Z_t.shape[0],))
prob_pred = self.iso.transform(Z_t)
return prob_pred
def predict(self, X_test):
if self.forcing == True:
X_test = X_test[:, :-1]
prob = self.predict_proba(X_test)
label_pred = np.asarray([1 if i >= 0.5 else 0 for i in prob])
return label_pred
Step 4: Visualizing result and Conclusion
def plot_reliability_diag():
calibrators = ['sigmoid', 'platt_scaling', 'isotonic']
fig = plt.figure(figsize=(12, 12))
ax1 = plt.subplot2grid((3, 1), (0, 0), rowspan=2)
ax2 = plt.subplot2grid((3, 1), (2, 0))
ax1.plot([0, 1], [0, 1], "k:", label="Perfectly calibrated")
for c in calibrators:
calibrator = CalibratedEstimator(svc, f'{c}').pre_train(X_train, y_train)
calibrator.fit(X_calib, y_calib)
y_pred = calibrator.predict_proba(X_test)
mean_predicted_value, fraction_of_positives = calibration_curve(y_test, y_pred, n_bins=10)
ax1.plot(mean_predicted_value, fraction_of_positives, "s-", label=f"SVC + {c}")
ax2.hist(y_pred, range=(0, 1), bins=10, label=f"SVC + {c}", histtype="step", lw=2)
platt_no_b = CalibratedEstimator(svc, 'platt_scaling', forcing=True).pre_train(X_train, y_train)
platt_no_b.fit(X_calib, y_calib)
y_pred_no_b = platt_no_b.predict_proba(X_test)
mean_predicted_value_no_b, fraction_of_positives_no_b = calibration_curve(y_test, y_pred_no_b, n_bins=10)
ax1.plot(mean_predicted_value_no_b, fraction_of_positives_no_b, "s-", label="SVC + platt(b=0)")
ax2.hist(y_pred_no_b, range=(0, 1), bins=10, label=f"SVC + platt(b=0)", histtype="step", lw=2)
ax1.set_ylabel("Fraction of positives", fontsize=16)
ax1.set_ylim([-0.05, 1.05])
ax1.legend(loc="lower right", fontsize=16)
ax1.set_title('Calibration plots (reliability curve)', fontsize=16)
ax2.set_xlabel("Mean predicted value", fontsize=16)
ax2.set_ylabel("Count", fontsize=16)
ax2.legend(loc="upper center", ncol=2, fontsize=16)
fig.savefig('calibration_result.png')
plot_reliability_diag()
output:
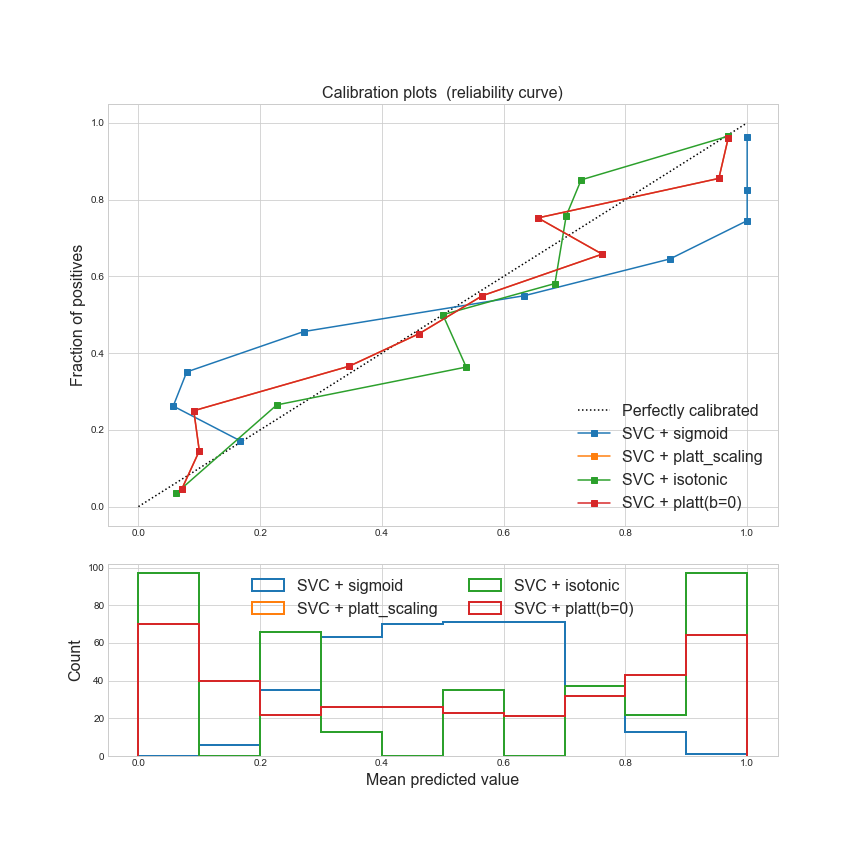
Conclusion
-
For
sigmoid function, calibration did not happen, since the squashing function just transforms real-valued scores \(z(x)\) to a range of \([0, 1]\). -
From the plot above, we conclude that the reliability curve for
platt scalingwith b overlaps the curve that forcing $b=0$, which indicates that the intercept does not affect the calibration result, meanwhile, platt scaling does a better calibration than option 1, as we can see its reliability curve is much closer to perfectly calibrated line. -
Compared to platt scaling, isotonic regression does not perform better here.
-
Whether to choose platt scaling or istonic regression, it depends on the situation, isotonic regression is suitable for a bigger calibration set.
-
For multiclass predictions, each class was calibrated separately in a OneVsRestClassifier fashion. When predicting probabilities, the calibrated probabilities for each class are predicted separately. As those probabilities do not necessarily sum to one, a postprocessing is performed to normalize them. References: sklearn calibration